Enlisting computational technologies in the war on cancer isn’t new but it has taken on an increasingly decisive role. At SC17, Eric Stahlberg, director of the HPC Initiative at Frederick National Laboratory for Cancer Research in the Data Science and Information Technology Program, and two colleagues will lead the third Computational Approaches for Cancer workshop being held the Friday, Nov. 17, at SC17.
It is hard to overstate the importance of computation in today’s pursuit of precision medicine. Given the diversity and size of datasets it’s also not surprising that the “new kids” on the HPC cancer fighting block – AI and deep learning/machine learning – are also becoming the big kids on the block promising to significantly accelerate efforts understand and integrate biomedical data to develop and inform new treatments.

In this Q&A, Stahlberg discusses the goals of the workshop, the growing importance of AI/deep learning in biomedical research, how programs such as the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) are progressing, the need for algorithm assurance and portability, as well as ongoing needs where HPC technology has perhaps fallen short. The co-organizers of the workshop include Patricia Kovatch, Associate Dean for Scientific Computing at the Icahn School of Medicine at Mount Sinai and the Co-Director for the Master of Science in Biomedical Informatics, and Thomas Barr, Imaging Manager, Biomedical Imaging Team, Research Institute at Nationwide Children’s Hospital.
HPCwire: Maybe set the framework of the workshop with an overview of its goals. What are you and your fellow participants trying to achieve and what will be the output?
Eric Stahlberg: Great question. Cancer is an extremely complex disease – with hundreds of distinct classifications. The scale of the challenge is such that it requires a team approach to make progress. Now in its third year, the workshop continues to provide a venue that brings together communities and individuals from all interests and backgrounds to work together to impact cancer with HPC and computational methods. The workshops continue to be organized to help share information and updates on new capabilities, technologies and opportunities involving computational approaches, with an eye on seeing new collaborative efforts develop around cancer.
The first year of the workshop in 2015 was somewhat remarkable. The HPC community attending SC has had a long history of supporting the cancer research community in many ways, yet an opportunity to bring the community together had not yet materialized. The original intent was simply to provide a venue for those with in interest in cancer to share ideas, bring focus to potential priorities and look ahead as to what might be possible. The timing was incredible, with the the launching of the National Strategic Computing Initiative in July, opening up a whole new realm of potential possibilities and ideas.
 By the time of the workshop last year, many of these possibilities started to gain traction – the Cancer Moonshot Initiative providing a strong and motivating context to get started, accelerate and make progress rapidly. Many new efforts were just getting off the ground, creating huge potential for collaboration – with established efforts employing computational approaches blending with new initiatives being launched as part of the Cancer Moonshot.
By the time of the workshop last year, many of these possibilities started to gain traction – the Cancer Moonshot Initiative providing a strong and motivating context to get started, accelerate and make progress rapidly. Many new efforts were just getting off the ground, creating huge potential for collaboration – with established efforts employing computational approaches blending with new initiatives being launched as part of the Cancer Moonshot.
The workshop this year continues to build on the direction and success of the first two workshops. Similar to the first two workshops, speakers are being invited to help inform on opportunities for large scale computing in cancer research. This year’s workshop will feature Dr. Shannon Hughes from the NCI Division of Cancer Biology delivering a keynote presentation highlighting many efforts at the NCI where HPC can make a difference, particularly in the area of cancer systems biology. In addition, this year’s workshop brings a special emphasis to the role of machine learning in cancer – a tremendously exciting area, while also providing an opportunity to update the HPC community on the progress of collaborative efforts of the NCI working with the Department of Energy.
The response to the call for papers this year was also very strong – reflecting the rapidly growing role of HPC in accelerating cancer research. In addition to a report to compile summaries the workshop contributions, highlight key issues, and identify new areas of exploration and collaboration, the organizers are anticipating a special journal issue where these contributions can also be shared in full.
HPCwire: In the fight against cancer, where has computational technology had the greatest impact so far and what kinds of computational infrastructure have been the key enablers? Where has its application been disappointing?
Stahlberg: Computational technology has been part of the cancer fight for many years, providing many meaningful contributions along the way to advance understanding, provide insight, and deliver new diagnostic capabilities. One can find computational technology at work in many areas including imaging systems, in computational models used in cancer drug discovery, in assembling, mapping and analyzing genomes that enable molecular level understanding, even in the information systems used to manage and share information about cancer patients.
Answering the question as to where has computational technology been most disappointing in the fight against cancer is also difficult to answer given the breadth of areas where computational technology has been employed. However, given the near-term critical need to enable greater access to clinical data including patient history and outcomes, an area where great promise remains is in the use of computational technologies that make it possible for more patient information to be brought together more fully, safely and securely to accelerate progress in the direction of clinical impact.
HPCwire: What are the key computational gaps in the fight against cancer today? How do you see them being addressed and what particular technologies, life science and computational, are expected to have the greatest impact?
Stahlberg: The computational landscape for cancer is changing so rapidly that the key gaps continue to also change. Not too long ago, there was great concern about the amount of data being generated and whether this data could all be used effectively. Within just a few years, the mindset has changed significantly, where the challenge is now focusing on bringing what data we have available together and recognizing that even then, the complexity of the disease demands even more [data] as we aim for more precision in diagnosis, prognosis, and associated treatments.
 With that said, computational gaps exist in nearly every domain and area of the cancer fight. At the clinical level, computation holds promise to help bring together, even virtually, the large amounts of existing data locked away in organizational silos. At the clinical level, there are important gaps in the data available for cancer patients pre and post treatment, a gap that may well be filled by both better data integration capabilities as well as mobile health monitoring. Understanding disease progression and response also presents a gap as well as an associated opportunity for computing and life science to work together to find efficient ways to monitor patient progress, track and monitor the disease at the cellular level, and create profiles of how different treatments impact the disease over time in the laboratory and in the clinic.
With that said, computational gaps exist in nearly every domain and area of the cancer fight. At the clinical level, computation holds promise to help bring together, even virtually, the large amounts of existing data locked away in organizational silos. At the clinical level, there are important gaps in the data available for cancer patients pre and post treatment, a gap that may well be filled by both better data integration capabilities as well as mobile health monitoring. Understanding disease progression and response also presents a gap as well as an associated opportunity for computing and life science to work together to find efficient ways to monitor patient progress, track and monitor the disease at the cellular level, and create profiles of how different treatments impact the disease over time in the laboratory and in the clinic.
It is clear that technologies in two areas will be key in the near term – machine learning and technologies that enable algorithm portability.
In the near term, machine learning is expected to play a very significant role, particularly as the amount of data generated is expected to grow. We have already seen progress in automating feature identification as well as delivering approaches for predictive models for complex data. As the amount of available, reliable cancer data across all scales increases, the opportunity for machine learning to accelerate insight and leverage these new levels of information will continue to grow tremendously.
A second area of impact, related to the first, is in technologies for algorithm assurance and portability. As computing technology has become increasingly integrated within instruments and diagnostics at the point of acquisition exploding the volume of data collected, the need grows tremendously to move algorithms closer to the point of acquisition to enable processing before transport. The need for consistency and repeatability in the scientific research process requires portability and assurance of the analysis workflows. Portability and assurance of implementation are also important keys to eventual success in a clinical setting.
Efforts in delivering portable workflows through containers are also demonstrating great promise in moving the compute to the data, are providing an initial means at overcoming existing organizational barriers to data access.
Extending a bit further, technologies that enable portability of algorithms and of trained predictive models will also become keys to future success for HPC in cancer research. As new ways to encapsulate knowledge in the form of a trained neural network, parameterized set of equations, or other forms of predictive models, having reliable, portable knowledge will be a key factor to share insight and build the collective body of knowledge needed to accelerate cancer research.
HPCwire: While we are talking about infrastructure could you provide a picture of the HPC resources that NIH/NCI have, are they sufficient as is, and what the plans are for expanding them?

Stahlberg: The HPC resource map for the NIH and NCI, like many large organizations, ranges from small servers to one of the largest systems created to support biological and health computation. There has been a wonderful recent growth in the available HPC resources available to NIH and NCI investigators, as part of a major NIH investment. The Biowulf team has done a fantastic job in raise the level of computing to now include 90,000+ processors. This configuration includes an expanded role of heterogeneous technologies in the form of GPUs, and presents a new level of computing capability available to the NIH and to NCI investigators.
In addition, NCI supports a large number of investigators through its multiple grant programs, where large scale HPC resources supported by NSF and others are being put to use in the war on cancer. While the specific details on the magnitude of computing this represents is not immediately available, the breadth of this level of support is expected to be quite substantial. At the annual meeting earlier this year for CASC (Coalition for Academic Scientific Computing), when asked which centers were supporting NCI funded investigators, nearly every attendee raised their hand.
Looking ahead, it would be difficult to make the case that even this level of HPC resources, will be sufficient as is, knowing the dramatic increases in the amount of data being generated currently, being forecast for the future, and the overall deepening complexity of cancer as new insights are revealed on an ongoing basis. With the emphasis on precision medicine and in the case of NCI, precision oncology, new opportunities to accelerate research and insight using HPC are quickly emerging.
Looking to the future of HPC and large scale computing in cancer research was one of the many factors supporting the new collaborative effort between the NCI and DOE. With the collaboration now having wrapped up the first year, new insights are being provided here that will merged with additional insights and information from the many existing efforts, to help inform future planning for HPC resources in the context of the emerging Exascale computing capabilities and emerging HPC technologies.
HPCwire: Interdisciplinary expertise is increasingly important in medicine. One persistent issue has been the relative lack of computational expertise among clinicians and life science researchers. To what extent is this changing and what steps are needed to raise computational expertise among this group?
Stahlberg: Medicine has long been a team effort, drawing from many disciplines and abilities to deliver care to the patient. There have been long-established centers in computational and mathematical aspects of medicine. One such example is the Advanced Biomedical Computing Center at the Frederick National Laboratory for cancer research which has been at the forefront of computational applications of medicine for more than twenty-five years. The difference today is the breadth of disciplines that are working together, and the depth of demand for computational scientists, as sources and volumes of available data in medicine and life sciences have exploded. The apparent shortage of computational expertise among clinicians and life sciences researchers is largely a result of the rapid rate of change in these areas, where the workforce and training have yet to catch up to the accelerating pace of technology and data-driven innovation.
Fortunately, many have recognized the need and opportunity for cross-disciplinary experience in computational and data sciences to enable ongoing advances in medicine. This appreciation has led to many new academic and training programs supported by NIH, NCI, as well as many catalyzed by health organizations and universities themselves that will help full future demand.
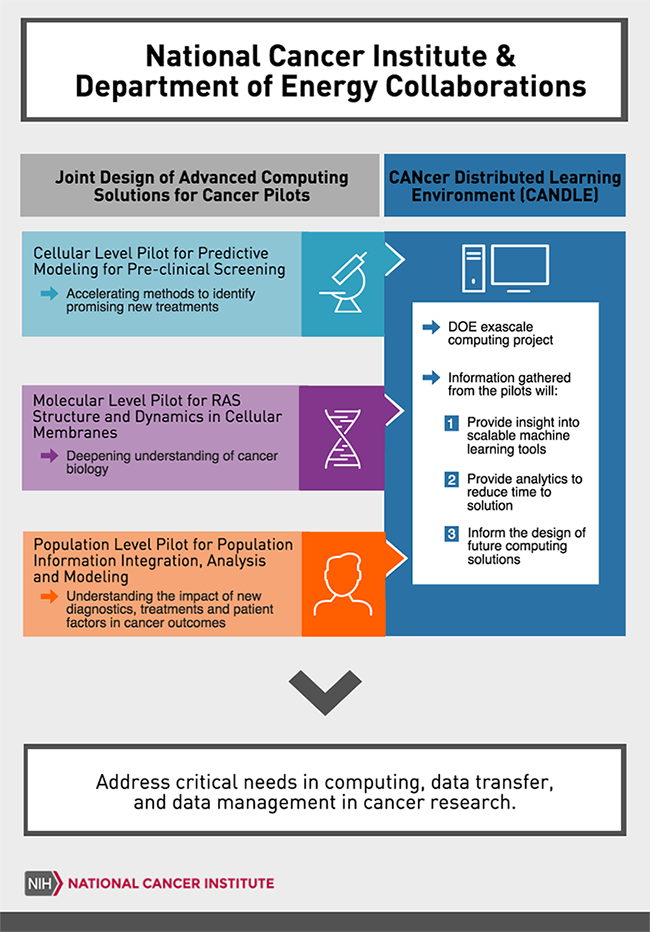
Collaborative opportunities between computational scientists and the medical research community are helping fill the immediate needs. One such example is the Joint Design of Advanced Computing Solutions for Cancer, a collaboration between the National Cancer Institute and the Department of Energy, brings together world-class computational scientists together with world-class cancer scientists in shared efforts to advance missions aims in both cancer and Exascale computing by pushing the limits of each together.
More organizations, seeing similar opportunities for cross-disciplinary collaboration in medicine, will certainly be needed to address the near-term demand while existing computational and data science programs adapt to embrace the medical and life sciences, and new programs begin to deliver the cross-trained, interdisciplinary workforce for the future.
HPCwire: Deep learning and Artificial Intelligence are the big buzzword in advanced scale computing today. Indeed, the CANDLE program efforts to learn how to apply deep learning in areas such as simulation (RAS effort), pre-clinical therapy evaluation, and outcome data mining are good examples. How do you see deep learning and AI being used near term and long-term in the war on cancer?
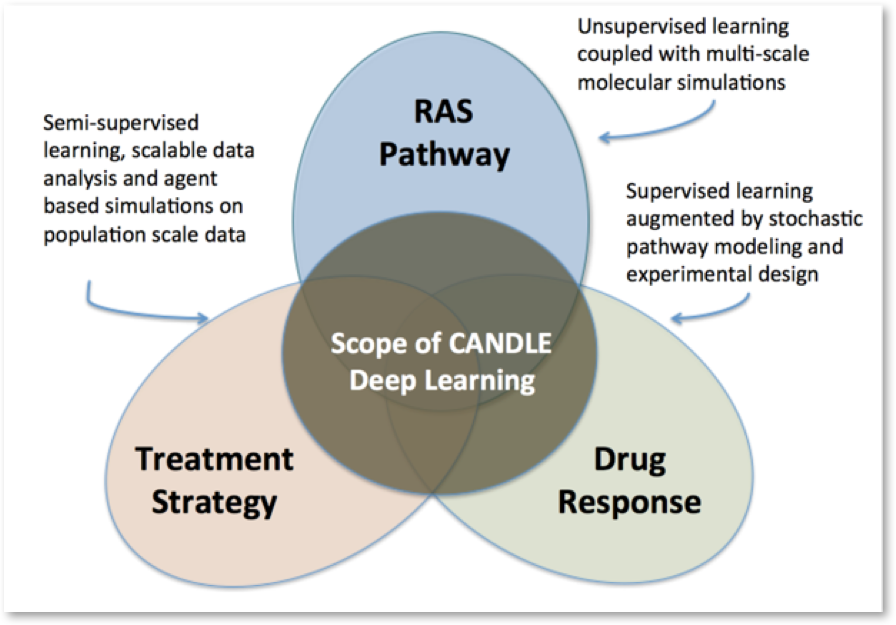 Stahlberg: While AI has a long history of application in medicine and life sciences, the opportunities for deep learning based AI in the war on cancer are just starting to be developed. As you mention, the application of deep learning in the JDACS4C pilots involving molecular simulation, pre-clinical treatment prediction, and outcome modeling are just developing the frontier of how this technology can be applied to effect and accelerate the war on cancer. The CANDLE Exascale Computing project, led by Argonne National Laboratory, was formed out of the recognition that AI and deep learning in particular was intrinsic to each pilot, and had broad potential application across the cancer research space. The specific areas being explored by the three pilot efforts as part of the CANDLE project provide some insight into how deep learning and AI can be expected to have future impact in the war on cancer.
Stahlberg: While AI has a long history of application in medicine and life sciences, the opportunities for deep learning based AI in the war on cancer are just starting to be developed. As you mention, the application of deep learning in the JDACS4C pilots involving molecular simulation, pre-clinical treatment prediction, and outcome modeling are just developing the frontier of how this technology can be applied to effect and accelerate the war on cancer. The CANDLE Exascale Computing project, led by Argonne National Laboratory, was formed out of the recognition that AI and deep learning in particular was intrinsic to each pilot, and had broad potential application across the cancer research space. The specific areas being explored by the three pilot efforts as part of the CANDLE project provide some insight into how deep learning and AI can be expected to have future impact in the war on cancer.
The pilot collaboration on cancer surveillance (pilot 3) led by investigators from the NCI Division of Cancer Control and Population Science and Oak Ridge National Laboratory demonstrating how deep learning can be applied to extract information from complex data, extracting biomarker information from electronic pathology reports. Similar capabilities have been shown to be possible with the processing of image information. Joined with automation, in the near term, deep learning can be expected to deepen and broaden the available insight about the cancer patient population in ways not otherwise possible.
The pilot collaboration on RAS-related cancers (pilot 2), led by investigators from Frederick National Laboratory and Lawrence Livermore National Laboratory follows in this direction, applying deep learning to extract and correlate features of potential interest from complex molecular interaction data.
The pilot collaboration on predictive cancer models (pilot 1), led by investigators from Frederick National Laboratory and Argonne National Laboratory are using deep learning based AI in a different manner, using deep learning to develop predictive models of tumor response. While still very early, the potential use of deep learning for the development of predictive models in cancer is very exciting, opening doors to many new avenues to develop a ‘cancer learning system’ that will join data, prediction, and feedback in a learning loop that holds potential to revolutionize how we prevent, detect, diagnose, and treat cancer.
In the era that combines scale of big data, exascale computing and deep learning, new levels of understanding about the data are also possible and extremely valuable. Led by scientists at Los Alamos National Laboratory, Uncertainty Quantification, or UQ, is also an important element of the NCI collaboration with the DOE. Providing key insights into limits of the data and limits of the models, the information provided by UQ is helping to inform new approaches and priorities to improve both the robustness of the data and the models being employed.
These are just a few of the near-term areas where deep learning and AI are anticipated to have an impact. Looking long-term, the role of these technologies is difficult to forecast, but in drawing parallels from other disciplines, some specific areas begin to emerge.
First, for making predictions on complex systems such as is with cancer, ensemble and multi-model approaches are likely to be increasingly required to build consensus among likely outcomes across a range of initial conditions and parameters. Deep learning is likely to be used in both representing the complex systems being modeled, but also to inform the selections and choices to be involved in the ensembles. In a second future scenario, data-driven deep learning models may also form a future basis for portably representing knowledge about cancer, particularly in recognition of the complexity of the data and ongoing need to maintain data provenance and security. Deep learning models may be readily developed with locally accessible datasets, then shared with the community without sharing the actual data.
As a third future scenario, in translation to critical use scenarios, as core algorithms for research or central applications in the clinic, deep learning models provide a means for maintaining consistency, validation and verification in translation from research to clinical setting.

HPCwire: One area that has perhaps been disappointing is predictive biology, and not just in cancer. Efforts to start with first principles, such as is done building modern jetliners, or even with experimental data from elucidation of various pathways, to ‘build’ drugs have had mixed results. Leaving aside things like structure scoring (docking, etc.), where is predictive biology headed in terms of fighting cancer and what’s the sense around needed technology requirements, for example specialized supercomputers such Anton1 and 2, and what’s the sense of needed basic knowledge to plug into predictive models?
Stahlberg: Biology has been a challenge for computational prediction given the overall complexity of the system and the role that subtle changes and effects can have in the overall outcome. The potential disappointment that may be assigned to predictive biology is most likely relative – relative to the what has been demonstrated in other disciplines such as transportation.
This is what makes the current era so very promising for accelerating progress on cancer and predictive biology. A sustained effort employing the lessons learned from other industries, where it is now increasingly possible to make the critical observations of biology at the fundamental level of the cell, combined with the computing capabilities that are rapidly becoming available, sets the stage for transforming predictive biology in a manner observed in parallel industries. Two elements of that transformation highlight the future direction.
First, the future for predictive biology is likely to be multi-scale, both in time and space, where models for subsystems are developed, integrated and accelerated computationally to support and inform predictions across multiple scales and unique biological environments, and ultimately for increasingly precise predictions for defined groups of individuals. Given the multi-scale nature of biology itself, the direction is not too surprising. The challenge is in getting there.
One of the compelling features for deep learning in the biological domain is in its flexibility and applicability across the range of scales of interest. While not a substitute for fundamental understanding, deep learning enables a first step to support a predictive perspective for the complexity of data available in biology. This first step enables active learning approaches, where data is used to develop predictive models that are progressively improved with new data and biological insight obtained from experimentation aimed at reducing uncertainty around the prediction.
A critical area of need already identified is the need more longitudinal observations and data with which to have both greater insight into outcomes for patients, but also greater insight into the incremental changes of biological state over time at all scales. In the near term, by starting with the data we currently have available, advances will be made to help inform on the data expected to be required for improved predictions, whereby insights will be gained to define the information truly needed for confident predictions.
The role of specialized technologies will be critical, particularly in the context of predictive models, as the size, complexity and number of subsystems are studied and explored to align predictions across scales. These specialized technologies will lead the forefront of efficient implementations of predictive models, increasing the speed and reducing the costs required to study and inform decisions for increasingly precise and predictive oncology.
Brief Bio:
Eric Stahlberg is director of the HPC Initiative at Frederick National Laboratory for Cancer Research in the Data Science and Information Technology Program. In this role he also leads HPC strategy and exploratory computing efforts for the National Cancer Institute Center for Biomedical Informatics and Information Technology (CBIIT). Dr. Stahlberg also spearheads collaborative efforts between the National Cancer Institute and the US Department of Energy in such efforts as that Joint Design for Advanced Computing Solutions for Cancer (JDACS4C), the CANcer Distributed Learning Environment (CANDLE), and Accelerating Therapeutics for Opportunities in Medicine (ATOM). Prior to joining Frederick National Laboratory, he directed an innovative undergraduate program in computational science, led efforts in workforce development, led HPC initiatives in bioinformatics, and multiple state and nationally funded projects.




























































