This blog post was contributed by Don Freed, Senior Bioinformatics Scientist, and Brendan Gallagher, Head of Business Development at Sentieon; and Olivia Choudhury, PhD, Senior Partner Solutions Architect, Sujaya Srinivasan, Genomics Solutions Architect, and Aniket Deshpande, Senior Specialist, HPC HCLS at AWS.
The year 2022 was an exciting one for genomics customers, with announcements from Illumina’s targeting $2/Gbase for 30X whole genomes, and new instruments from Element Biosciences, Ultima Genomics, and Pacific Biosciences. While these platforms offer exciting new choices for genomic data generation, they also present new challenges for data analysis due to unique platform-specific data characteristics and the large amount of data generated. These challenges highlight the need for efficient and scalable solutions for genomic data processing that are flexible enough to handle the unique properties of these emerging platforms.
Sentieon develops highly-optimized algorithms for bioinformatics applications, providing accurate, efficient, performant, and robust tools for the analysis of next generation sequencing data. Sentieon DNAseq and TNseq provide an implementation of the GATK best practices pipelines for germline and somatic variant calling with improved robustness and performance. Sentieon’s DNAscope pipelines are shown to be highly performant on Illumina data while offering substantially improved germline variant calling accuracy compared to GATK best practices.
In this blog post, we benchmark the performance of Sentieon’s DNAseq and DNAscope pipelines using publicly available datasets from Illumina, PacBio HiFi, Element Biosciences, and Ultima Genomics platforms using Amazon Elastic Compute Cloud (Amazon EC2) instances. Readers will gain an understanding of the runtime, cost, and accuracy performance of state-of-the-art germline variant calling pipelines across a wide range of Amazon EC2 instances. These benchmarks use publicly available datasets and the pipeline is available on GitHub.
Benchmarking Sentieon’s DNAseq and DNAscope pipelines
Pre-requisites
- AWS account with permission to provision Amazon EC2, Amazon EBS, and Amazon VPC.
- Deployment of a Sentieon license server inside of your VPC. Sentieon provides a Deployment Guide for AWS at, https://support.sentieon.com/appnotes/aws_deployment/.
- An EC2 instance within the same VPC as the Sentieon license server.
Running Sentieon on AWS
An implementation of the Sentieon pipeline used in this blog post along with scripts for instance setup are provided on GitHub. The command `bash instance_setup.sh` will download the appropriate build of the Sentieon software and install other dependencies for this benchmark workflow. The pipelines benchmarked in this blog can be run with `bash run_benchmarks.sh`. The pipelines will download the input datasets, reference, and model files, down-sample the input read data to a consistent number of bases, run the Sentieon secondary analysis pipelines with the input dataset, and calculate variant calling accuracy against the Genome in a Bottle (GIAB) v4.2.1 truth set.
In practice, we ran the first part of the pipeline once to prepare the files used in the benchmark and then stored the prepared files on an AWS FSx for Lustre file system. The preprocessed files were used in later benchmarks on other instance types.
An Amazon gp3 EBS volume with 10,000 IOPS and a throughput of 700 MB/s was used for intermediate file storage. We found that this EBS disk worked well with the demanding IO needs of the Sentieon pipelines on the largest instances. For benchmarks using the hpc6a.48xlarge instance, the EBS disk was supplemented with a ramdisk for faster IO operations.
Benchmark Analysis
In these benchmarks, we use the GIAB HG002 sample sequenced on multiple sequencing platforms. As described in the pipeline config file, input FASTQ files were obtained from the following URLs:
- Illumina NovaSeq
- https://s3.amazonaws.com/genomics-benchmark-datasets/google-brain/fastq/novaseq/wgs_pcr_free/40x/HG002.novaseq.pcr-free.40x.R1.fastq.gz
- https://s3.amazonaws.com/genomics-benchmark-datasets/google-brain/fastq/novaseq/wgs_pcr_free/40x/HG002.novaseq.pcr-free.40x.R2.fastq.gz
- Illumina HiSeq
- https://s3.amazonaws.com/genomics-benchmark-datasets/google-brain/fastq/novaseq/wgs_pcr_free/40x/HG002.novaseq.pcr-free.40x.R1.fastq.gz
- https://s3.amazonaws.com/genomics-benchmark-datasets/google-brain/fastq/novaseq/wgs_pcr_free/40x/HG002.novaseq.pcr-free.40x.R2.fastq.gz
- Element Biosciences Aviti
- Downloaded from https://go.elementbiosciences.com/access-seq-datasets-060622
- Ultima Genomics UG100
- https://s3.amazonaws.com/ultima-selected-1k-genomes/crams/005401-UGAv3-1-CACATCCTGCATGTGAT.cram
- PacBio HiFi
- https://s3.amazonaws.com/giab/data/AshkenazimTrio/HG002_NA24385_son
/PacBio_CCS_15kb_20kb_chemistry2/reads/m64011_190830
_220126.fastq.gz - https://s3.amazonaws.com/giab/data/AshkenazimTrio/HG002_NA24385_son
/PacBio_CCS_15kb_20kb_chemistry2/reads/m64011_190901
_095311.fastq.gz - https://s3.amazonaws.com/giab/data/AshkenazimTrio/HG002_NA24385_son
/PacBio_CCS_15kb_20kb_chemistry2/reads/m64012_190920
_173625.fastq.gz - https://s3.amazonaws.com/giab/data/AshkenazimTrio/HG002_NA24385_son
/PacBio_CCS_15kb_20kb_chemistry2/reads/m64012_190921
_234837.fastq.gz - https://s3.amazonaws.com/giab/data/AshkenazimTrio/HG002_NA24385_son
/PacBio_CCS_15kb_20kb_chemistry2/reads/m64015_190920
_185703.fastq.gz - https://s3.amazonaws.com/giab/data/AshkenazimTrio/HG002_NA24385_son
/PacBio_CCS_15kb_20kb_chemistry2/reads/m64015_190922
_010918.fastq.gz
- https://s3.amazonaws.com/giab/data/AshkenazimTrio/HG002_NA24385_son
The input files vary in their coverage, so the datasets with FASTQ input were down-sampled to approximately 93 billion bases (~30x coverage) prior to processing with the Sentieon secondary analysis pipelines. The Ultima CRAM file was not down-sampled and is at 40x coverage as recommended by Ultima Genomics.
The data were processed using the hg38 reference genome. The reference genome at https://giab.s3.amazonaws.com/release/references/GRCh38/GCA_000001405.15
_GRCh38_no_alt_analysis_set.fasta.gz was used for files with input in the FASTQ format. The reference genome at https://broad-references.s3.amazonaws.com/hg38/v0/Homo_sapiens_assembly38.fasta was used with the Ultima data in CRAM format, as this dataset was already aligned to this reference genome.
Benchmarking Sentieon’s variant calling pipelines with Hpc6a instances
The Sentieon software is highly flexible, and can be used to process data with a wide variety of EC2 instance types. The recently announced Hpc6a instance family offers competitive price performance for compute intensive genomic secondary analysis workloads and is a good fit for Sentieon’s secondary analysis pipelines. Figure 1 presents the runtime and On-Demand compute cost of running Sentieon’s secondary analysis pipelines for germline variant calling across multiple sequencing technologies on hpc6a.48xlarge instances in the US East (Ohio) Region at the time of publication.

On the hpc6a.48xlarge instance, analyzing a 30x Illumina NovaSeq sample from FASTQ to VCF took Sentieon’s DNAseq pipeline data 32 minutes with an On-Demand compute cost of $1.53. Sentieon’s DNAscope pipeline reduces SNP errors by 53% and INDEL errors by 78% on the 30x Illumina NovaSeq dataset compared to DNAseq while taking only 3-5 minutes longer — about 35 to 38 minutes for the various short-read platforms. On-Demand cost for DNAseq and DNAscope pipelines was typically $1.75, give or take a few cents.
The Ultima UG100 dataset was already aligned to the reference genome and in this benchmark, we only perform variant calling without alignment. Variant calling (CRAM to VCF) with the Ultima dataset finished in under 22 minutes for $1.05 in On-Demand compute costs on hpc6a.48xlarge.
Sentieon’s DNAscope LongRead pipeline for PacBio HiFi data is more computationally intensive as it includes multiple passes of variant calling along with a read-backed phasing. The DNAscope LongRead pipeline finished in 77 minutes with an On-Demand cost of $3.71.
The Element Biosciences AVITI system is a new desktop sequencer that became commercially available in the Spring of 2022 and is supported by a customized Sentieon DNAscope pipeline. Sentieon’s DNAscope pipeline for Element Biosciences finished in 35 minutes with an On-Demand compute cost of $1.69.
A full breakdown of the run times and costs are in Table 1.
Table 1: Runtime and On-Demand compute cost of the stages of the Sentieon DNAseq and DNAscope pipelines. Alignment includes alignment with Sentieon BWA-MEM for short-read data and alignment with Sentieon minimap2 for PacBio HiFi data. Preprocessing includes duplicate marking, base-quality score recalibration, and merging of multiple aligned files into a single file. Variant calling includes variant calling or variant candidate identification along with variant genotyping and filtering. Variant calling for PacBio HiFi data is implemented as a multi-stage pipeline. All analysis were done on hpc6a.48xlarge instances in the US East (Ohio) Region and are accurate at the time of publishing this post.
| Sample | Pipeline | Alignment (min) | Preprocessing (min) | Variant Calling (min) | Total Runtime (min) | On-Demand hourly rate ($/hr) | On-Demand compute cost ($) |
| Element Aviti | DNAscope | 23.2 | 1.1 | 10.8 | 35.2 | 2.88 | 1.69 |
| Illumina HiSeq X | DNAseq | 23.7 | 2.6 | 4.2 | 30.5 | 2.88 | 1.46 |
| Illumina HiSeq X | DNAscope | 23.7 | 1.0 | 10.5 | 35.2 | 2.88 | 1.69 |
| Illumina NovaSeq | DNAseq | 25.1 | 2.3 | 4.5 | 31.9 | 2.88 | 1.53 |
| Illumina NovaSeq | DNAscope | 25.1 | 0.9 | 11.6 | 37.6 | 2.88 | 1.81 |
| PacBio HiFi | DNAscope | 24.8 | 1.6 | 50.9 | 77.2 | 2.88 | 3.71 |
| Ultima UG100 | DNAscope | N/A | N/A | 21.2 | 21.2 | 2.88 | 1.02 |
Variant calling accuracy of the Sentieon DNAseq and DNAscope pipelines
The variant calling accuracy of the Sentieon pipelines as measured against the GIAB v4.2.1 truthset for the HG002 sample is shown in Figure 2. Sentieon’s DNAscope provides highly accurate SNP and indel variant calling across all platforms, with SNP F1-scores exceeding 99.5% and indel F1-scores exceeding 99.2%. Sentieon’s DNAseq pipeline provides identical results to the GATK best practices pipeline for germline variant calling, but has lower accuracy on the GIAB benchmark dataset.

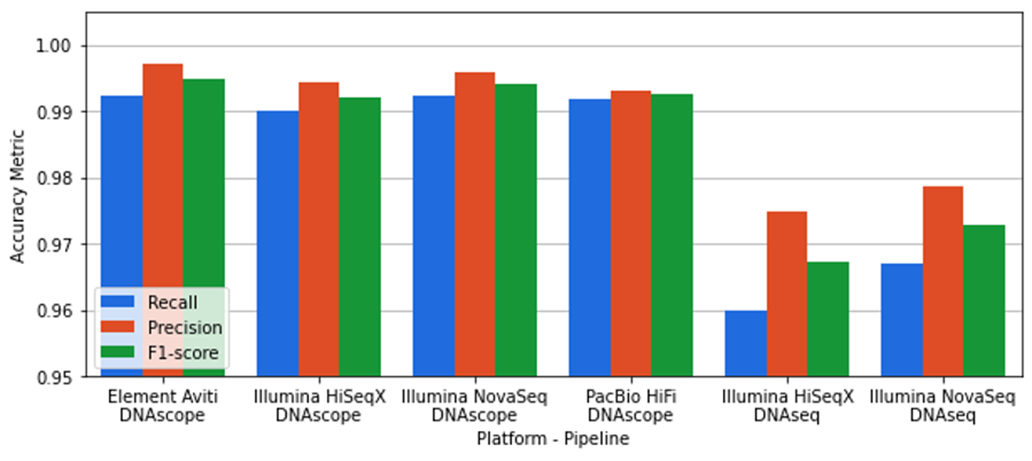
The Ultima Genomics UG100 platform generates 20-24 WGS samples per day with a single instrument using a novel sequencing chemistry. During each sequencing cycle, this chemistry may incorporate zero, one, or more bases and the measured signal strength is proportional to the number of incorporated bases. Ultima Genomics, in their initial publication with the Broad Institute, has defined a high-confidence region which excludes 1.8% of the GIAB v4.2.1 high-confidence region with long homopolymers and other difficult regions…
Read the full blog to learn more. Reminder: You can learn a lot from AWS HPC engineers by subscribing to the HPC Tech Short YouTube channel, and following the AWS HPC Blog channel.