A new record for HPC scaling on the public cloud has been achieved on Microsoft Azure. Led by Dr. Jer-Ming Chia, the cloud provider partnered with the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign (UIUC) to test the limits of Azure’s scalability on a key molecular dynamics tool. The result: successful scaling across a staggering 86,400 CPU cores — a virtual cluster with nearly 5 peak petaflops of performance.
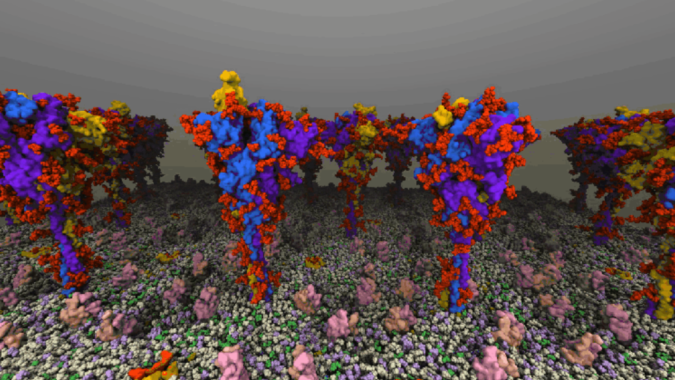
The code at hand was Nanoscale Molecular Dynamics (NAMD), developed in-house at UIUC. NAMD, nicknamed “the computational microscope,” has played a major role in COVID-19 research this year and was at the heart of the massive simulations of the coronavirus that yesterday won the 2020 ACM Gordon Bell Special Prize in HPC-Based COVID-19 Research. Azure in fact hosted similar simulations of SARS-CoV-2’s viral envelope and spike proteins as part of a project funded by the COVID-19 HPC Consortium.
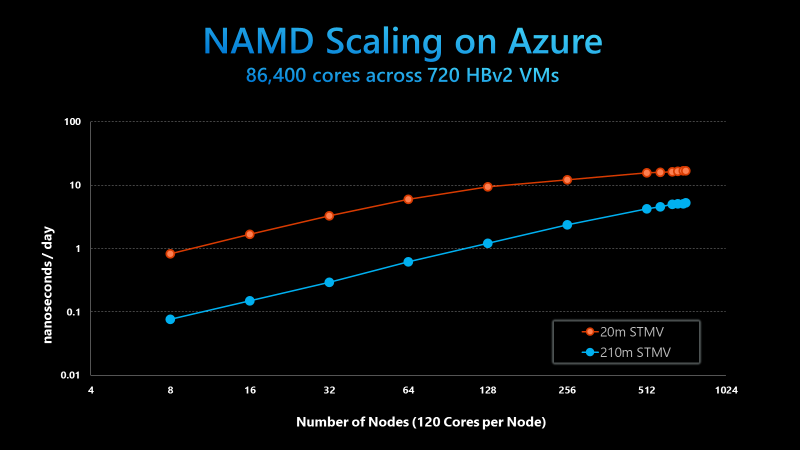
The efforts focused on scaling Azure’s HBv2 virtual machines, each of which was equipped with 120 AMD Epyc 7002-series CPU cores, 480GB of memory and – crucially – 200 Gbps Nvidia HDR InfiniBand. Using the HBv2 VMs, the team scaled NAMD on 20 million-atom and 210 million-atom systems. The 210 million-atom system scaled up to 720 nodes, achieving what Azure claims is the largest HPC simulation ever on the public cloud by an order of magnitude.
“The team found not only could HBv2 clusters meet the researchers’ requirements, but that performance and scalability on Azure rivaled and in some cases surpassed the capabilities of the Frontera supercomputer, ranked 8th on the June 2020 Top500 list,” wrote Evan Burness, principal program manager for Azure HPC.
The successful scaling bodes well for future molecular dynamics projects in the cloud, as the test was intended to evaluate HBv2 VMs for future SARS-CoV-2 simulations.
“Our NIH center has put in years of programming effort towards developing fast scalable codes in NAMD,” said David Hardy, a senior research programmer at UIUC. “These efforts along with the UCX [communication layer] support in Charm++ [the library NAMD uses for parallelization] allowed us to get good performance and scaling for NAMD on the Microsoft Azure Rome cluster without any other modifications to the source code.”
Looking ahead, Azure will likely begin to deploy new hardware soon. Recently, the cloud provider announced that it would be using next-generation AMD Epyc CPUs – codenamed “Milan” – to power future HB-series virtual machines for HPC use cases. AMD is currently preparing to ship the Milan-series CPUs.
Header image: a still from a NAMD simulation of SARS-CoV-2. Image courtesy of UIUC.